Guide RNA structure design enables combinatorial CRISPRa programs for biosynthetic profiling
Published in Nature Communications, 2024
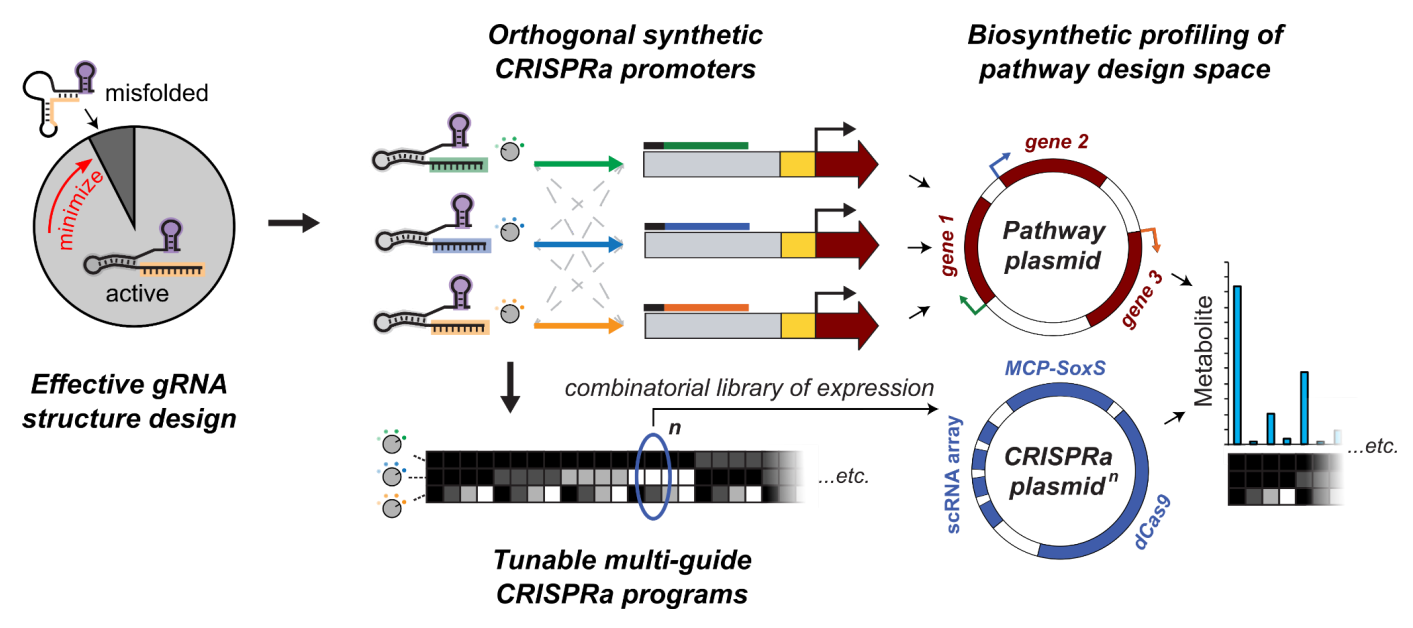
Abstract
Engineering bacterial metabolism to efficiently produce chemicals and materials from multi-step pathways requires optimizing multi-gene expression programs to achieve enzyme balance. CRISPR-Cas transcriptional control systems are emerging as important metabolic engineering tools for programming multi-gene expression regulation. However, poor predictability of guide RNA folding can disrupt enzyme balance through unreliable expression control. We devised a set of computational parameters that can describe guide RNA folding, and we expect them to be broadly applicable across CRISPR-Cas9 systems. Here, we correlate efficacy of modified guide RNAs (scRNAs) for CRISPR activation (CRISPRa) in E. coli with a kinetic parameter describing folding rate into the active structure. This parameter also enables forward design of new scRNAs, with no observed failures in our screen. We use CRISPRa target sequences from this set to design a system of three synthetic promoters that can orthogonally activate and tune expression of chosen outputs over a >35-fold dynamic range. Independent activation tuning allows experimental exploration of a three-dimensional expression design space via a 64-member combinatorial triple-scRNA library. We apply these CRISPRa programs to two biosynthetic pathways, demonstrating production of valuable pteridine and human milk oligosaccharide products in E. coli. Profiling these design spaces indicated expression combinations producing up to 2.3-fold higher titer than that produced by maximal expression. Mapping production can also identify bottlenecks as targets for pathway redesign, improving titer of the oligosaccharide lacto-N-tetraose by 6-fold. Aided by computational scRNA efficacy prediction, the combinatorial CRISPRa strategy enables effective optimization of multi-step metabolic pathways. More broadly, the guide RNA design rules uncovered here may enable the routine design of effective multi-guide programs for a wide range of model- and data-driven applications of CRISPR gene regulation in bacterial hosts.
Recommended citation: Fontana J.+, Sparkman-Yager D.+, Faulkner I.+, Cardiff R., Kiattisewee C., Walls A., Primo T.G., Kinnunen P.C., Garcia Martin H., Zalatan J.G.†, Carothers J.M.† (2024). "Guide RNA structure design enables combinatorial CRISPRa programs for biosynthetic profiling." Nature Communications. 15:6341. PMID: 39068154
